Complete genomic profiles of 1496 Taiwanese reveal curated medical insights
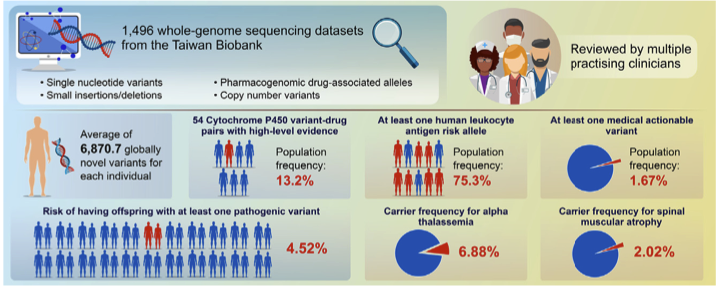
We found that each participant had an average of 6,870.7 globally novel variants and 75.3% (831/1103) of the participants harbored at least one PharmGKB-selected high evidence level human leukocyte antigen (HLA) risk allele. 54 PharmGKB-reported high-level instances of evidence of Cytochrome P450 variant-drug pairs, with a population frequency of over 13.2%. We also identified 23 variants in the ACMG secondary finding V3 gene list from 25 participants, suggesting that 1.67% (25/1496) of the population is harboring at least one medical actionable variant. Our carrier status analyses suggest that one in 25 couples (3.94%) would risk having offspring with at least one pathogenic variant, which is in line with rates found in Japan and Singapore. For pathogenic CNV, we detected 6.88% and 2.02% carrier rates for alpha thalassemia and spinal muscular atrophy, respectively.
< Link >
AI for Life Science and Precision Medicine
An AutoML solution for Epigenomics analysis
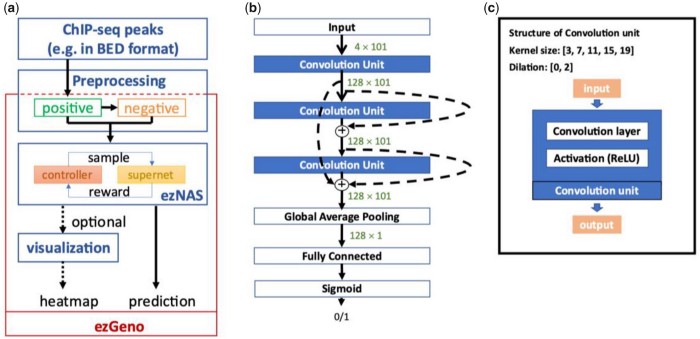
To facilitate the process of tailor-making a deep neural network for exploring the dynamics of genomic DNA, we have developed a hands-on package called ezGeno. ezGeno automates the search process of various parameters and network structures and can be applied to any kind of 1D genomic data. Combinations of multiple abovementioned 1D features are also applicable. The ezGeno package can be freely accessed at
https://github.com/ailabstw/ezGeno.
< Link >
DockCoV2: a drug database against SARS-CoV-2
We built DockCoV2 to provide the binding affinity of FDA-approved and Taiwan National Health Insurance (NHI) drugs with seven proteins. This database contains a total of 3,109 drugs. DockCoV2 is available at https://covirus.cc/drugs/
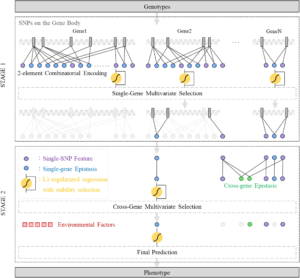
GenEpi: gene-based epistasis discovery using machine learning
We developed an efficient and effective GWAS method to detect epistasis for discovering sophisticated pathogenesis, which is especially important for complex diseases such as Alzheimer's disease (AD). GenEpi is a computational package to uncover epistasis associated with phenotypes by the proposed machine learning approach. GenEpi identifies both within-gene and cross-gene epistasis through a two-stage modeling workflow. In both stages, GenEpi adopts two-element combinatorial encoding when producing features and constructs the prediction models by L1-regularized regression with stability selection. The simulated data showed that GenEpi outperforms other widely-used methods on detecting the ground-truth epistasis. As real data is concerned, this study uses AD as an example to reveal the capability of GenEpi in finding disease-related variants and variant interactions that show both biological meanings and predictive power.
< Link >
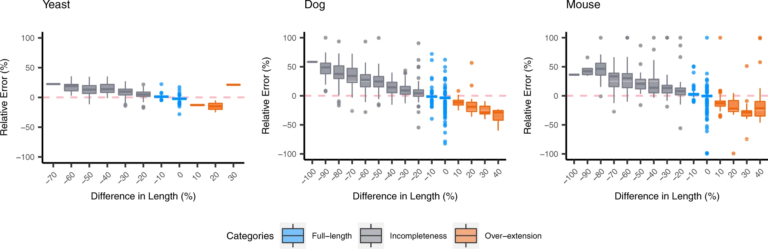
Effect of de novo transcriptome assembly on transcript quantification
For the organisms without the reference transcriptome, de novo transcriptome assembly must be carried out prior to quantification. This study investigates how assembly quality affects the performance of quantification based on de novo transcriptome assembly. We examined the over-extended and incomplete contigs, and demonstrated that assembly completeness has a strong impact on the estimation of contig abundance. Then we investigated the behavior of the quantifiers with respect to sequence ambiguity which might be originally presented in the transcriptome or accidentally produced by assemblers. The results suggested that the quantifiers often over-estimate the expression of family-collapse contigs and under-estimate the expression of duplicated contigs. For organisms without reference transcriptome, it remains challenging to detect the inaccurate estimation on family-collapse contigs. On the contrary, we observed that the situation of under-estimation on duplicated contigs can be warned through analyzing the read proportion of estimated abundance (RPEA) of contigs in the connected component inferenced by the quantifiers. In addition, we suggest that the estimated quantification results on the connected component level have better accuracy over sequence level quantification.
< Link >
Whole-genome de novo sequencing reveals unique genes that contributed to the adaptive evolution of the Mikado pheasant
攝影:謝郁震
The Mikado pheasant 帝雉 (Syrmaticus mikado) is a nearly endangered species indigenous to high-altitude regions of Taiwan. We completed the draft genome of the Mikado pheasant, which consists of 1.04 Gb of DNA and 15,972 annotated protein-coding genes. The Mikado pheasant displays expansion and positive selection of genes related to features that contribute to its adaptive evolution, such as energy metabolism, oxygen transport, hemoglobin binding, radiation response, immune response, and DNA repair. To investigate the molecular evolution of the major histocompatibility complex (MHC) across several avian species, 39 putative genes spanning 227 kb on a contiguous region were annotated and manually curated. The MHC loci of the pheasant revealed a high level of synteny, several rapidly evolving genes, and inverse regions compared to the same loci in the chicken. The complete mitochondrial genome was also sequenced, assembled, and compared against four long-tailed pheasants. The results from molecular clock analysis suggest that ancestors of the Mikado pheasant migrated from the north to Taiwan about 3.47 million years ago.
< Link >
[Dosudo] DeepLearning MEET #11 陳倩瑜教授專訪
隨著基因定序技術的成熟及便利, 基因組資料已經變成深度學習下一個最受矚目及最具潛力的焦點, 各大公司也紛紛佈局在AI醫療及基因研究上. 這次 Dosudo 的專訪榮幸的邀請到臺大生物產業機電工程學系的陳倩瑜教授來為我們談談如何將深度學習技術應用在基因資料的分析, 以及為何生物資訊需要結合人工智慧及這領域未來的展望.
< Link >
Integrating RNA-seq and ChIP-seq Data to Characterize Long Non-coding RNAs in Drosophila melanogaster
Recent advances in sequencing technology have opened a new era in RNA studies. Novel types of RNAs such as long non-coding RNAs (lncRNAs) have been discovered by transcriptomic sequencing and some lncRNAs have been found to play essential roles in biological processes. However, only limited information is available for lncRNAs in Drosophila melanogaster, an important model organism. Therefore, the characterization of lncRNAs and identification of new lncRNAs in D. melanogaster is an important area of research. Moreover, there is an increasing interest in the use of ChIP-seq data (H3K4me3, H3K36me3 and Pol II) to detect signatures of active transcription for reported lncRNAs.
In this study, we have developed a computational approach to identify new lncRNAs from two tissue-specific RNA-seq datasets using the poly(A)-enriched and the ribo-zero method, respectively. In our results, we identified 462 novel lncRNA transcripts, which we combined with 4137 previously published lncRNA transcripts into a curated dataset. We then utilized 61 RNA-seq and 32 ChIP-seq datasets to improve the annotation of the curated lncRNAs with regards to transcriptional direction, exon regions, classification, expression in the brain, possession of a poly(A) tail, and presence of conventional chromatin signatures. Furthermore, we used 30 time-course RNA-seq datasets and 32 ChIP-seq datasets to investigate whether the lncRNAs reported by RNA-seq have active transcription signatures. The results showed that more than half of the reported lncRNAs did not have chromatin signatures related to active transcription. To clarify this issue, we conducted RT-qPCR experiments and found that ~95.24 % of the selected lncRNAs were truly transcribed, regardless of whether they were associated with active chromatin signatures or not.
< Link >
Discovery of Genes Related to Formothion Resistance in Oriental Fruit Fly (Bactrocera dorsalis) by a Constrained Functional Genomics Analysis
Oriental fruit fly is a very destructive pest of fruit in areas where it occurs and some of them have insecticide resistance.
The underlying basis of such phenomena can involve complex interactions of multiple genes. This study aims to analyze the specific genes are involved in resistance mechanisms.
< Link >
De novo transcriptome sequencing of axolotl blastema for identification of differentially expressed genes during limb regeneration
It's amazing for salamanders are unique among vertebrates in their ability to completely regenerate amputated limbs through the mediation of blastema cells located at the stump ends. This study aims to know the secret in transcription mechanism of salamander's regeneration.
< Link >
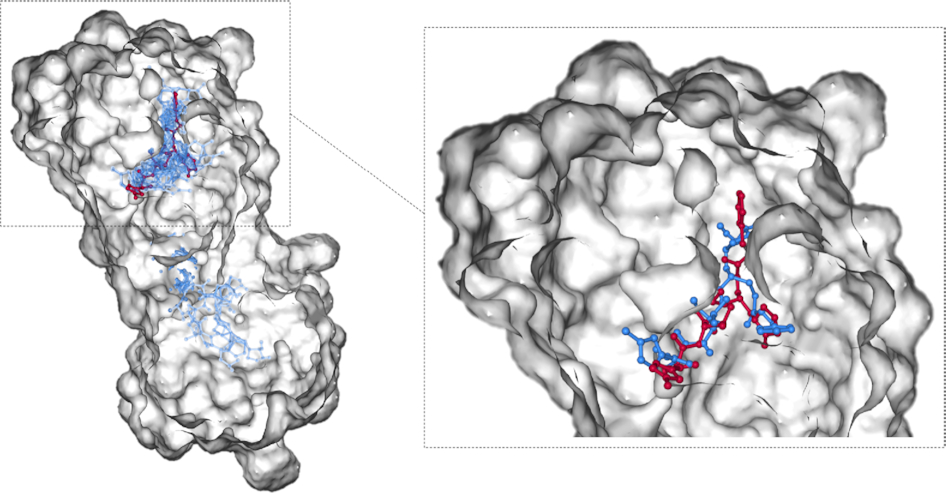
PiDNA: predicting protein-DNA interactions with structural models
Predicting binding sites of a transcription factor in the genome is an important, but challenging, issue in studying gene regulation. PiDNA, aims at first constructing reliable PWMs by applying an atomic-level knowledge-based scoring function on numerous in silico mutated complex structures, and then using the PWM constructed by the structure models with small energy changes to predict the interaction between proteins and DNA sequences. This study first shows that the constructed PWMs are similar to the annotated PWMs collected from databases or literature. Second, the prediction accuracy of PiDNA in detecting relatively high-specificity sites is evaluated by comparing the ranked lists against in vitro experiments from protein-binding microarrays. Finally, PiDNA is shown to be able to select the experimentally validated binding sites from 10 000 random sites with high accuracy.
PiDNA is available at:
here.
< Link >
Discovery of genes related to insecticide resistance in Bactrocera dorsalis by functional genomic analysis of a de novo assembled transcriptome
Insecticide resistance has recently become a critical concern for control of many insect pest species. Genome sequencing and global quantization of gene expression through analysis of the transcriptome can provide useful information relevant to this challenging problem. This study aims at using whole transcriptome analysis developed through de novo assembly to know resistance mechanisms from molecular aspect.
< Link >
De novo motif discovery facilitates identification of interactions between transcription factors in Saccharomyces cerevisiae
Previous studies have used target genes shared by two TFs as a clue to infer TF-TF interactions. However, the target genes with low binding affinity are frequently omitted by experimental data, especially when a single strict threshold is employed. This study aims at improving the accuracy of inferring TF-TF interactions.
< Link >
Predicting Target DNA Sequences of DNA-binding Proteins Based on Unbound Structures
DNA-binding proteins such as transcription factors use DNA-binding domains (DBDs) to bind to specific sequences in the genome to initiate many important biological functions. This study aims at investigating the possibility of predicting the DNA sequences bound by DNA-binding proteins from the proteins' unbound structures (structures of the unbound state).
< Link >
WildSpan: Mining Structured Motifs from Protein Sequences
Automatic extraction of motifs from biological sequences is an important research problem in study of molecular biology. For proteins, it is desired to discover sequence motifs containing a large number of wildcard symbols, as the residues associated with functional sites are usually largely separated in sequences. This paper proposes an algorithm named WildSpan (sequential pattern mining across large wildcard regions) that incorporates several pruning strategies to largely reduce the mining cost.
< Link >
Discovering gapped binding sites of yeast transcription factors
We propose a method for discovering TFBSs, especially gapped motifs. We use ChIP-chip data to judge the binding strength of a TF to a putative target promoter and use orthologous sequences from related species to judge the degree of evolutionary conservation of a predicted TFBS.
< Link >
Protein Disorder Prediction by Condensed PSSM Considering Propensity for Order or Disorder
More and more disordered regions have been discovered in protein sequences, and many of them are found to be functionally significant. Previous studies reveal that disordered regions of a protein can be predicted by its primary structure, the amino acid sequence.
Recent studies further show that employing evolutionary information such as position specific scoring matrices (PSSMs) improves the prediction accuracy of protein disorder. As more and more machine learning techniques have been introduced to protein disorder detection, extracting more useful features with biological insights attracts more attention.
< Link >
c4Lab was founded
c4Lab was founded in the department of Biomechartronics Engineering, National Taiwan University by Dr. Chien-Yu Chen and her students.
Incremental Generation of Summarized Clustering Hierarchy for Protein Family Analysis
Protein sequence clustering has been widely exploited to facilitate in-depth analysis of protein functions and families. For some applications of protein sequence clustering, it is highly desirable that a hierarchical structure, also referred to as dendrogram, which shows how proteins are clustered at various levels, is generated. In this paper, the design of a novel incremental clustering algorithm aimed at generating summarized dendrograms for analysis of protein databases is described. The proposed incremental clustering algorithm employs a statistics-based model to summarize the distributions of the similarity scores among the proteins in the database and to control formation of clusters.
< Link >